
The nucleosomes.ĭuring replication, two nucleosomes in front of the replication fork, that is unreplicated DNA, becomes destabilized. Histone H1 forms the linker between two nucleosomes. Each nucleosome is composed of 8 histone proteins, two each of H2A, H2B, H3 and H4.
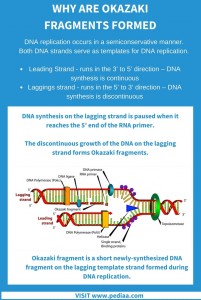
It involves winding of the negatively charged DNA around the basic proteins called histones to form structures called nucleosomes (fig below). As we know, one of the feature of the eukaryotic DNA is that it is packaged in form of highly compact structures called the Chromosomes. Fig 4: Removal of RNA primer and linking of individual Okazaki fragments.ĭuring elongation, there is continuous disassembly as reassembly of the nucleosome packaging along the DNA, too. The nick formed between the Okazaki fragment and the lagging strand are filled in by DNA ligase I, hence forming single lagging strand. Polymerase δ then fills the gaps formed between Okazaki fragments following the primer removal. This left out ribonucleotide is then removed by flap endonuclease 1 ( FEN 1). RNA primers are removed by RNase H1, such that one ribonucleotide remains still attached to the DNA (3′ end) part of the Okazaki fragment. ( Just for info: Know more about PCNA) The proteins involved in the replication, especially PCNA (Boehm et al., 2016). DNA pol δ dissociates after the synthesis of the entire DNA stretch, as it approaches the previous RNA primer. Following which, the RFC causes polymerase switching and the deoxyribonucleotides are added by DNA pol δ held by the PCNA, the sliding clamp (see the figure below). Hence an Okazaki fragment is made up of RNA nucleotides (7-10 nucleotides), then around 10-20 nucleotides of DNA bases are added by the DNA pol α. As several Okazaki fragments are made, Polymerase switching during synthesis of Okazaki fragments is of higher importance. Okazaki fragments (fig 3) generated in eukaryotes during lagging-strand synthesis are around 200 bases (prokaryotes, around 2000 bases) long. The replication in the lagging strands is discontinuous and takes place with formation of several Okazaki fragments (fig 2). Polymerases δ is the major polymerase in lagging-strand synthesis.
#Difference between okazaki fragment and lagging strand series
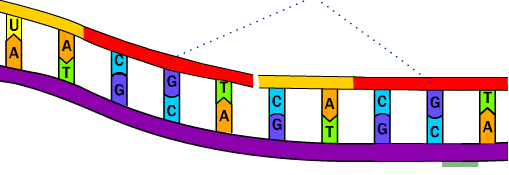
Lagging strand synthesis: Fig 2: Synthesis of lagging or discontinuous strand with series of Okazaki fragments.

In the leading strand as the replication is continuous and the primer is synthesized only once and the extension is carried out (fig 1). Pol ε has been reported as the main leading strand synthesis polymerase (in Saccharomyces cerevisiae). Leading strand synthesis: Fig 1: Synthesis of leading or continuous strand.Īs DNA pol α completes synthesizing RNA primer and adding DNA bases, the RFC causes dissociation of DNA pol α and assembles proliferating cell nuclear antigen (PCNA) in the region of the primer terminus. (Please read about the lagging strand, leading strands and the Okazaki fragments in a previous post.)
The DNA strands are then extended by other polymerases namely the DNA polymerase δ and DNA polymerase ε. Replication factor C (RFC) initiates a reaction called polymerase switching. The primase subunit synthesizes the RNA primers (around 7-12 nucleotides) which are then transferred to the polymerase domain and extended with DNA bases (around 20-25 nucleotides). The first polymerase to initiate the DNA synthesis is the DNA polymerase α, which exists in the form of DNA polymerase α-primase complex. ( Just for info: Read more about the eukaryotic origins in the paper titled ‘ Making Sense of Eukaryotic DNA Replication Origins‘.) Elongation: The polymerases work together with other proteins for the elongation of the daughter strands. To this protein complex the polymerases are loaded. In the present post, let’s look into the Elongation and the Termination in Eukaryotic DNA Replication.ĭuring Initiation, a repertoire of proteins bind and unwind the DNA at the origin. We discussed about the Initiation in the Eukaryotic cells in the last post.


 0 kommentar(er)
0 kommentar(er)
